Principal Component Analysis in R
I wanted to know a little more on Principal Component Analysis (PCA) in R. For this purpose, I first created my own artificial dataset. I wanted to reuse the same dataset later on for performing also cluster analysis, so I put a little bit of thought in how to create it.
This is the R code I used.
Classes <- sample(1:3, 100, replace = TRUE)
createData <- function(class, means1, sd1, means2, sd2) {
n <- length(class)
X1 <- rnorm(n, means1[class], sd1)
X2 <- rnorm(n, means2[class], sd2)
X3 <- sample(-100:100, n, replace=TRUE)
X4 <- X1 + X1 * runif(n, -0.8, 0.8)
X5 <- rnorm(n, -18, 1.2)
m <- matrix(c(X3, X1, X4, X2, X5), nrow = length(X1), ncol = 5)
colnames(m) <- c("X3", "X1", "X4", "X2", "X5")
return(m)
}
data <- createData(Classes, c(3, 50, 100), 5, c(0.3, 0.5, 1.0), 0.1)Let’s go through this step by step.
First, I created a vector of length 100 containing values 1, 2 or 3: Classes <- sample(1:3, 100, replace = TRUE). The value would indicate later whether a certain observation in my dataset belongs to one of three different clusters of data.
Next, I created a function that can be used to create a dataset automatically. The created dataset has five different variables X1 to X5. X3 is a uniformly distributed random variable in the range of -100 to 100: X3 <- sample(-100:100, n, replace=TRUE). X5 is normally distributed random variable with mean -18 and a standard deviation of 1.2: X5 <- rnorm(n, -18, 1.2). In other words, X3 and X5 are totally unrelated to all other variables. In contrast, X1, X2 and X4 are all correlated to each other. An observation is placed in one of three different clusters of observations along three dimensions denoted as X1, X2 and X4. X1 has three cluster centers at 3, 50 and 100 and a standard deviation of 5: X1 <- rnorm(n, means1[class], sd1); X2 has three cluster centers at 0.3, 0.5 and 1.0 and a standard deviation of 0.1: X2 <- rnorm(n, means2[class], sd2); X4 is derived from X1: X4 <- X1 + X1 * runif(n, -0.8, 0.8). Note that I reordered all variables somewhat to make our task a little juicier.
Here’s the correlation matrix and the correlogram.
cor(data)
pairs(data)| X3 | X1 | X4 | X2 | X5 | |
| X3 | 1.00000000 | -0.1133516 | -0.08752043 | -0.09320869 | -0.02701275 |
| X1 | -0.11335158 | 1.0000000 | 0.83200505 | 0.90965123 | -0.14800759 |
| X4 | -0.08752043 | 0.8320051 | 1.00000000 | 0.75789451 | -0.15882644 |
| X2 | -0.09320869 | 0.9096512 | 0.75789451 | 1.00000000 | -0.13261305 |
| X5 | -0.02701275 | -0.1480076 | -0.15882644 | -0.13261305 | 1.00000000 |
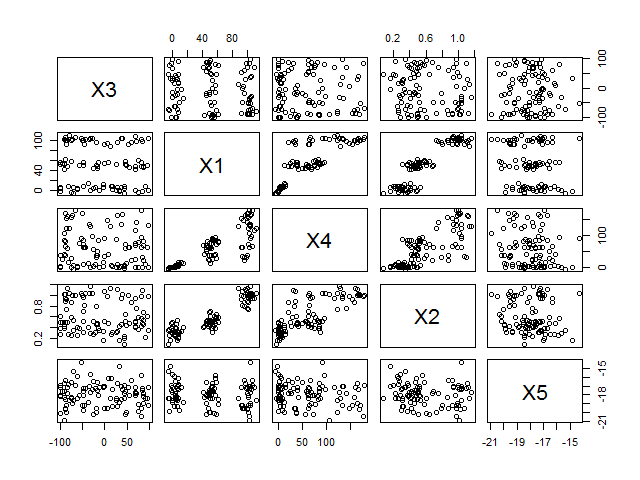
It should already be obvious from both the correlation matrix and the correlogram that there is a strong connection between X1, X2 and X4, whereas X3 and X5 are unrelated to the rest. Let’s see whether we can confirm this with a PCA. We use the princomp function with the default parameters.
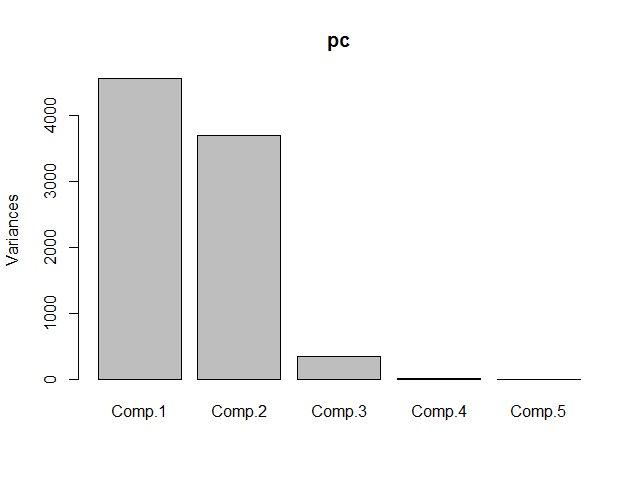
pc <- princomp(data)
summary(pc)
plot(pc)| Importance of components: | Comp.1 | Comp.2 | Comp.3 | Comp.4 | Comp.5 |
|---|---|---|---|---|---|
| Standard deviation | 67.5243440 | 60.8182793 | 18.54506263 | 1.2692048845 | 1.216295e-01 |
| Proportion of Variance | 0.5299356 | 0.4299031 | 0.03997229 | 0.0001872259 | 1.719413e-06 |
| Cumulative Proportion | 0.5299356 | 0.9598388 | 0.99981105 | 0.9999982806 | 1.000000e+00 |
The table shows that already component 1 and 2 together capture 96% of the data’s variance. The screeplot confirms this.
To find out which components match to which variables, we’ll use the loadings function.
loadings(pc)| Loadings | Comp.1 | Comp.2 | Comp.3 | Comp.4 | Comp.5 | |
|---|---|---|---|---|---|---|
| X3 | 0.596 | 0.803 | ||||
| X1 | -0.459 | 0.322 | 0.828 | |||
| X4 | -0.659 | 0.501 | -0.560 | |||
| X2 | 1.000 | |||||
| X5 | 1.000 |
| Comp.1 | Comp.2 | Comp.3 | Comp.4 | Comp.5 | |
|---|---|---|---|---|---|
| SS loadings | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Proporation Var | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 |
| Cumulative Var | 0.2 | 0.4 | 0.6 | 0.8 | 1.0 |
princomp by default uses the variance matrix - not the correlation matrix - to compute the different components. Variances unlike correlations, however, depend strongly on the scale and units of the input data. This means, either __[we must scale our input data prior to performing a principal component analysis or we must base our analysis on the correlations, not the variances, between our variables](http://stats.stackexchange.com/questions/53/pca-on-correlation-or-covariance)__.
Let's quickly have a look at the variance matrix.
var(data)| X3 | X1 | X4 | X2 | X5 | |
| X3 | 4043.545354 | -287.916544 | -307.35406 | -1.74489036 | -2.22236234 |
| X1 | -287.916544 | 1595.569658 | 1835.40627 | 10.69703439 | -7.64903703 |
| X4 | -307.354064 | 1835.406265 | 3049.97811 | 12.32218479 | -11.34843658 |
| X2 | -1.744890 | 10.697034 | 12.32218 | 0.08666848 | -0.05051051 |
| X5 | -2.222362 | -7.649037 | -11.34844 | -0.05051051 | 1.67390100 |
cor = TRUE parameter in the princomp function.
pc <- princomp(data, cor = TRUE)
summary(pc)
plot(pc)| Importance of components: | Comp.1 | Comp.2 | Comp.3 | Comp.4 | Comp.5 |
|---|---|---|---|---|---|
| Standard deviation | 1.649629 | 1.013166 | 0.9597193 | 0.50290695 | 0.27972026 |
| Proportion of Variance | 0.544255 | 0.205301 | 0.1842122 | 0.05058308 | 0.01564869 |
| Cumulative Proportion | 0.544255 | 0.749556 | 0.9337682 | 0.98435131 | 1.00000000 |
loadings(pc)| Loadings | Comp.1 | Comp.2 | Comp.3 | Comp.4 | Comp.5 |
|---|---|---|---|---|---|
| X3 | 0.772 | 0.628 | |||
| X1 | -0.586 | -0.201 | 0.779 | ||
| X4 | -0.551 | 0.801 | -0.218 | ||
| X2 | -0.569 | 0.118 | -0.564 | -0.587 | |
| X5 | 0.144 | -0.635 | 0.759 |
| </td> | Comp.1 | Comp.2 | Comp.3 | Comp.4 | Comp.5 | </tr> |
|---|---|---|---|---|---|---|
| SS loadings | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| Proportion Var | 0.2 | 0.2 | 0.2 | 0.2 | 0.2 | |
| Cumulative Var | 0.2 | 0.4 | 0.6 | 0.8 | 1.0 |
